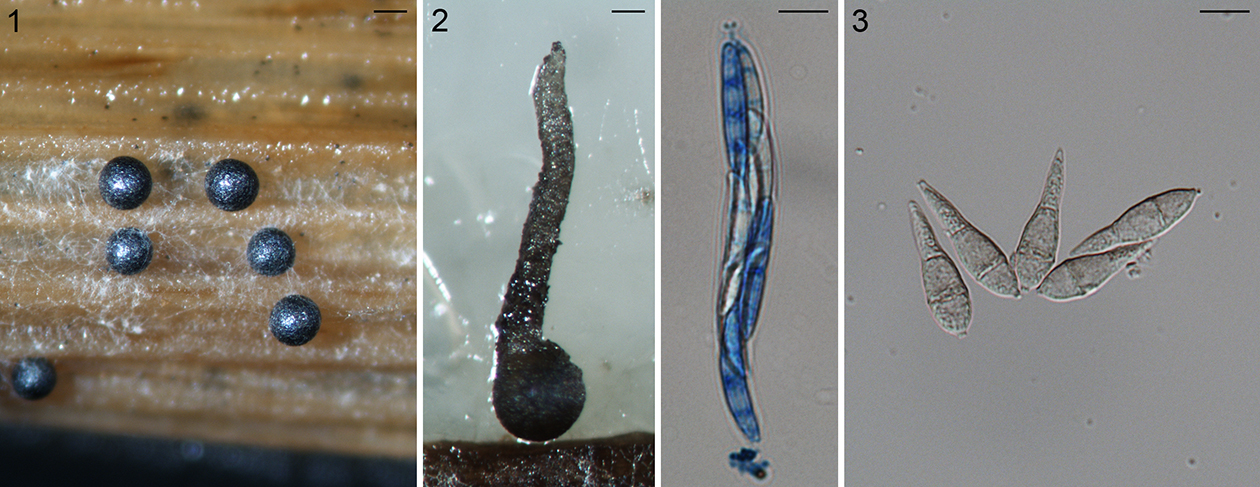
Fig. 1.1. Sclerotia of the rice stem rot fungus Nakataea oryzae, scale bar = 200 µm. 1.2. Perithecium (left, scale bar = 100 µm), ascus and ascospores (right, scale bar = 10 µm) of Magnaporthiopsis poae. 1.3. Pear-shaped conidia of Pyricularia grisea, scale bar = 10 µm.
The first described species in Magnaporthales was the rice stem rot fungus. The stem rot fungus, as Sclerotium oryzae, based on the sclerotial morph was named in 1877 (Cattaneo, 1877) (Fig. 1.1). Its conidial morphs were subsequently named Nakataea sigmoidea in 1939 (Hara, 1939). The sexual morph of this fungus was not discovered until after almost a century later. Krause and Webster (1972) established a new genus Magnaporthe in Diaporthales to accommodate the sexual morph of this species as Magnaporthe salvinii (Krause and Webster, 1972). Four more species have been placed in Magnaporthe based on their sexual morphology: M. grisea, M. oryzae, M. poae, and M. rhizophila. (Fig. 1.2).
In 1880, Saccardo established the genus Pyricularia (pyri: pear-shaped conidia, Fig. 1.3) based on P. grisea on crabgrass (Saccardo, 1880) including primarily asexual species. The rice isolates were designated as P. oryzae by Cavara (1892), which now is known as the rice blast fungus. Since then, over 70 names have been placed in Pyricularia many of which cause blast diseases of monocotyledonous plants. However, only a fraction of Pyricularia species have been included in modern phylogenetic analyses (Tosa and Chuma, 2014).
The sexual morph of P. grisea was first developed from crossing experiments in culture by Hebert (Hebert, 1971) and it was placed in Ceratosphaeria. Barr (1977) later moved it to Magnaporthe because of the similarity in ascospore morphology to the type species M. salvinii. The sexual morph of the rice blast fungus also was obtained by laboratory crossing experiments and was named M. oryzae (Couch and Kohn, 2002). Recent phylogenetic and phylogenomic analyses demonstrate that the rice blast fungus does not belong to Magnaporthe (Luo et al., 2015a; Luo and Zhang, 2013; Murata et al., 2014; Zhang et al., 2011) (Fig. 1.4). Rather the type of Magnaporthe, M. salvinii, belongs in Magnaporthaceae and is not congeneric with P. grisea and P. oryzae placed in Pyriculariaceae. Placement of the sexual morph of the rice blast fungus, P. oryzae, in Magnaporthe was based on incomplete data.
Traditionally, mycologists have given considerable weight to ascospore morphology, such as shape, size, pigmentation, and wall ornamentation, in delimiting genera in ascomycetes. With the development of polymerase chain reaction (PCR) and DNA sequencing technology in the early 1990s, molecular phylogeny has become a powerful approach for understanding fungal systematics. Molecular phylogenetic studies suggest that taxonomy based on traditional morphological characteristics does not always reflect evolutionary history. A 6-gene phylogeny, specifically the largest subunit of RNA polymerase II gene (RPB1), translation elongation factor 1-alpha gene (TEF1), a DNA replication licensing factor gene (MCM7), the internal transcribed spacer region of the nuclear rRNA genes (ITS), 18S rRNA gene (18S), and 28S rRNA gene (28S), has been applied to genera in Magnaporthales. Zhang et al. (2011) pointed out that both Gaeumannomyces and Magnaporthe, two important genera in Magnaporthales, are polyphyletic. Taxonomic revisions of these taxa subsequently have been made by Luo and Zhang (2013), Klaubauf et al. (2014), Hernández-Restrepo et al. (2016) and Zhang et al. (2016). Pathology and characteristics of the asexual morph, rather than ascospore morphology, correspond better with phylogenetic relationships of these fungi. The 6-gene phylogeny also recognizes two major clades in these fungi, one clade includes root-infecting species that produce phialophora-like conidia, while the other clade includes fungi causing blast diseases that produce pyricularia-like conidia (Zhang et al., 2011).
The family Magnaporthaceae was proposed for a group of cereal and grass-associated fungi centered on species causing rice stem rot and blast symptoms, along with Buergenerula, Clasterosphaeria, Gaeumannomyces, Herbampulla, and Omnidemptus, most of which are necrotrophic or hemibiotrophic pathogens (Cannon 1994). Early taxonomic studies had placed the rice blast fungus and allied species in different orders in Ascomycota, such as Diaporthales, Phyllachorales, and Xylariales (Barr, 1977; Krause and Webster, 1972). Later molecular phylogenetic analyses supported a monophyletic Magnaporthaceae as defined by (Cannon, 1994) placing it in subclass Sordariomycetidae (Sordariomycetes, Ascomycota). However, subsequent analyses supported variable phylogenetic affinities for this family. For example, in Zhang and Blackwell’s (2001) 18S rDNA tree, Magnaporthaceae formed a sister clade with Ophiostomatales. The 28S rDNA tree generated by Huhndorf et al. (2008) proposed a close relationship of Magnaporthaceae with Sordariales, Chaetosphaeriales and Boliniales (Huhndorf et al., 2008). In the 18S tree of Thongkantha et al. (2009), the Magnaporthaceae formed a clade sister to Diaporthales and Ophiostomatales; however, in the same paper, the 28S rDNA tree grouped them with Chaetosphaeriales and Sordariaceae (Thongkantha et al., 2009). Zhang et al. (2006) generated a four-gene phylogeny (TEF1; the second largest subunit of RNA polymerase II, RPB2; 18S and 28S) and suggested that the rice blast fungus was most closely related to Diaporthales. However, no other taxa in Magnaporthaceae were included in the analysis (Zhang et al., 2006). Poor taxon sampling due to a lack of sequence data from non-model species in Magnaporthaceae and phylogenetic heterogeneity among gene loci (Ebersberger et al., 2012; Fitzpatrick et al., 2006) likely explain the conflicting tree topologies in these studies.
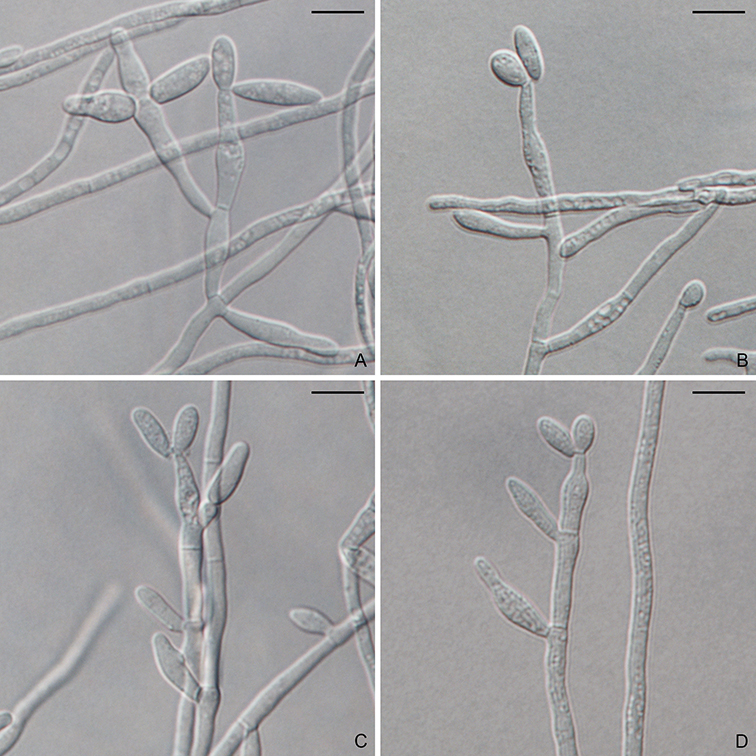
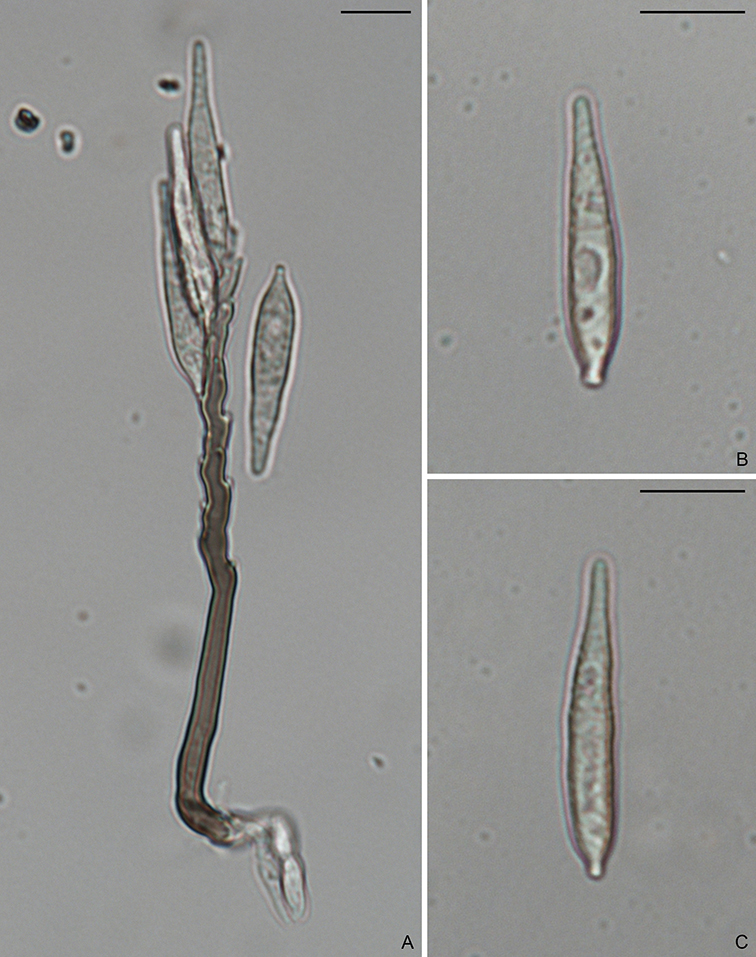
In 2009, a new order, Magnaporthales, with a single family was established (Thongkantha et al., 2009), characterized by nonstromatic black perithecia, usually with long hairy necks, persistent asci, and fusiform (spindle-shaped) or filiform (needle-shaped) ascospores. The asexual morphs are hyphomycetous and variable but can be categorized as two types: pyricularia-like or phialophora-like (Fig. 1.4). Several asexual genera have been linked to Magnaporthales, including Clasterosporium, Didymobotryum, Harpophora, Mycoleptodiscus, Nakataea, Phialophora, Pseudotracylla, Pyricularia, Sclerotium, and Trichocladium (Cannon, 1994; Cannon and Alcorn, 1994; Gams, 2000; Huhndorf et al., 2008; Kohlmeyer and Volkmann-Kohlmeyer, 1995; Thongkantha et al., 2009). Currently Clasterosporium, Nakataea, Pseudotracylla, and Pyricularia are maintained in Magnaporthales (Luo and Zhang, 2013; Zhang et al., 2016). Harpophora is treated as a synonym of Gaeumannomyces (Luo et al., 2015b) while Didymobotryum, Mycoleptodiscus, Phialophora, Sclerotium and Trichocladium are excluded from Magnaporthales (Klaubauf et al., 2014). Some pathogenic species produce specialized infection structures when colonizing their host plants, known as appressoria and hyphopodia. The hyphal pressing organs can generate strong turgor pressure to penetrate the host cells (Dean et al., 2005). Magnaporthales currently contains over 200 species. In addition, an increasing number of environmental sequences of uncultured Magnaporthales have been deposited in GenBank, suggesting cryptic diversity of Magnaporthales.
To generate a robust phylogeny for Magnaporthales, Luo et al. (2015a) performed a phylogenomic analysis using more than 200 genes, which supported Magnaporthales as a monophyletic order, sister to Ophiostomatales (Fig. 1.5) (Luo et al., 2015a). This result is consistent with the shared characteristics between the two orders: both have non-stromatic perithecia and a hyphomycetous asexual morph. In contrast, species in Diaporthales typically produce stromatic perithecia and a coelomycetous asexual morph. Within Magnaporthales, the phylogenomic analysis resulted in three well-supported clades: 1) an early diverging clade A comprised of saprotrophs associated with submerged wood; 2) clade B that includes the rice blast fungus and other pathogens that cause blast diseases of monocot plants. These species infect the above-ground tissues of host plants using a penetration structure, the appressorium; and 3) clade C comprised primarily of root-associated species that penetrate the root tissue with hyphopodia (Fig. 1.4). These three clades correspond to the families Magnaporthaceae, Ophioceraceae, and Pyriculariaceae as classified by Klaubauf et al. (2014) based on a 2-locus (28S and RPB1) phylogeny (Klaubauf et al., 2014). The early diverging lineage of Magnaporthales, namely Ophioceraceae (clade A) is mostly aquatic and saprotrophic on wood substrates. The pathogenicity of the remaining lineages is likely a derived feature from Pyriculariaceae (clade B), which evolved to infect host aerial parts using appressoria, and Magnaporthaceae (clade C) adapted to plant root infection with hyphopodia (Luo et al., 2015a). These well-supported phylogenies provide a robust framework for elucidating evolution of pathogenesis, nutrition modes, and phenotypic characters in Magnaporthales.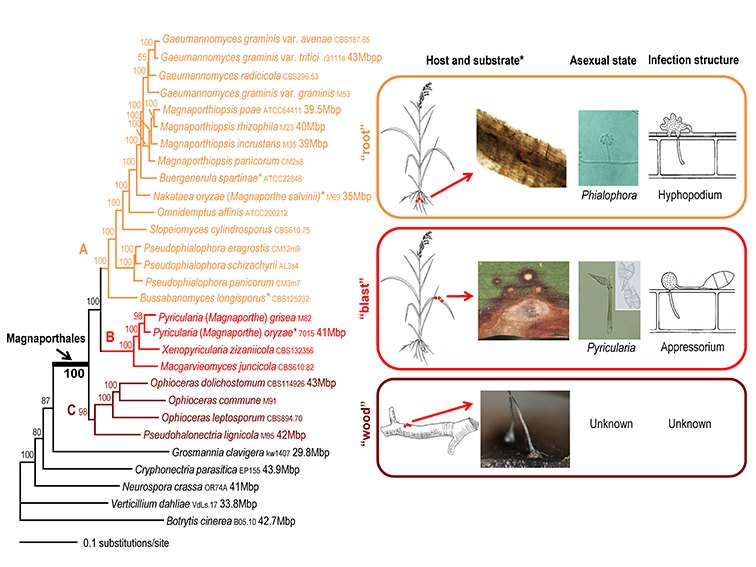
Fig. 1.4. Maximum likelihood tree of 24 Magnaporthales species and five Sordariomycetes used as outgroup species based on 82,715 amino acid positions derived from 226 genes. The infection sites and the morphology of sexual and asexual states for the three major Magnaporthales clades are illustrated (right panel). The supporting values for each node were estimated using 1000 bootstrap replicates. The strain number and genome size (if available) for each species are provided (Luo et al. 2015a).
*Buergenerula spartinae, Bussabanomyces longisporus, Nakataea oryzae, and Omnidemptus affinis are associated with above-ground parts of host plants, which is an exception in clade C. Pyricularia oryzae is associated with both the leaf and root of the host plant, which is an exception in clade B.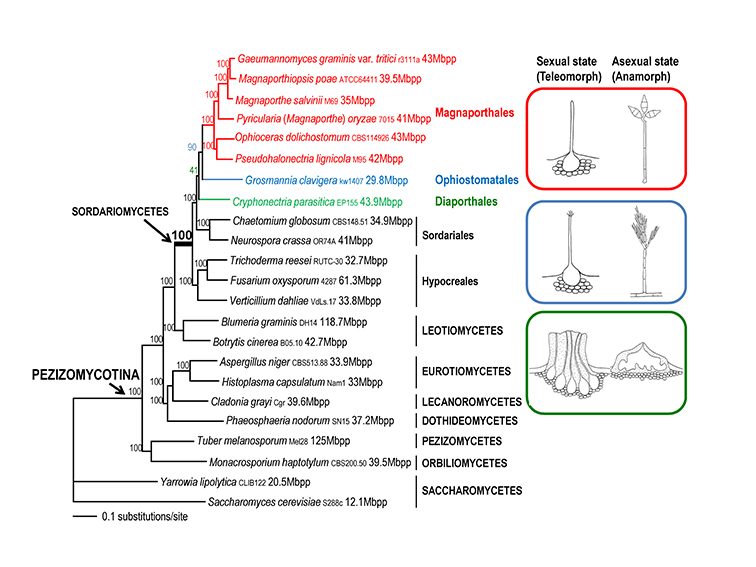
Fig. 1.5. Maximum likelihood tree of 21 Pezizomycotina species and two Saccharomycetes used as outgroup species based on 83,616 amino acid positions derived from 226 genes. The morphology of the sexual and asexual states for Magnaporthales and closely related species is shown (right panel). The supporting values for each node were estimated using 1000 bootstrap replicates. The strain number and genome size for each species are provided (Luo et al. 2015a).
In this monograph, we analyzed a number of DNA sequence data for several understudied taxa of Magnaporthales, such as Ceratosphaerella, Muraeriata, and Tropohalonectria in order to resolve their phylogenetic positions.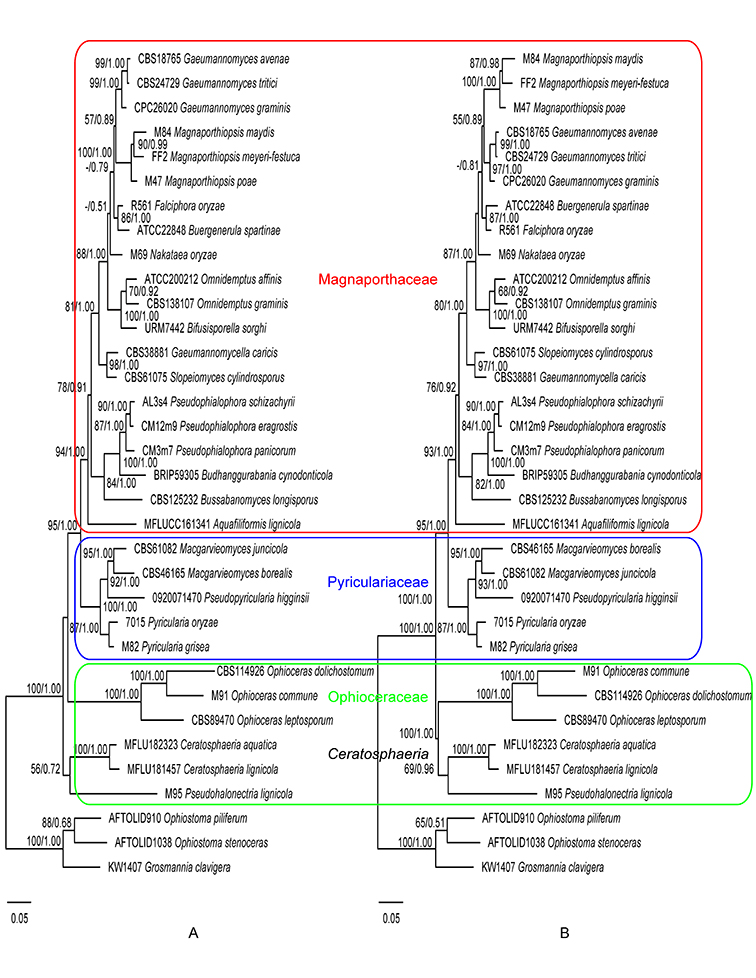
Fig. 1.6. Maximum likelihood trees inferred from the combined ITS, 28S, and TEF1 gene sequences without constraint (A), and with Magnaporthaceae, Pyriculariaceae, and Ophioceraceae as constraints (B). ML bootstrap values and BI posterior probabilities are indicated above internodes.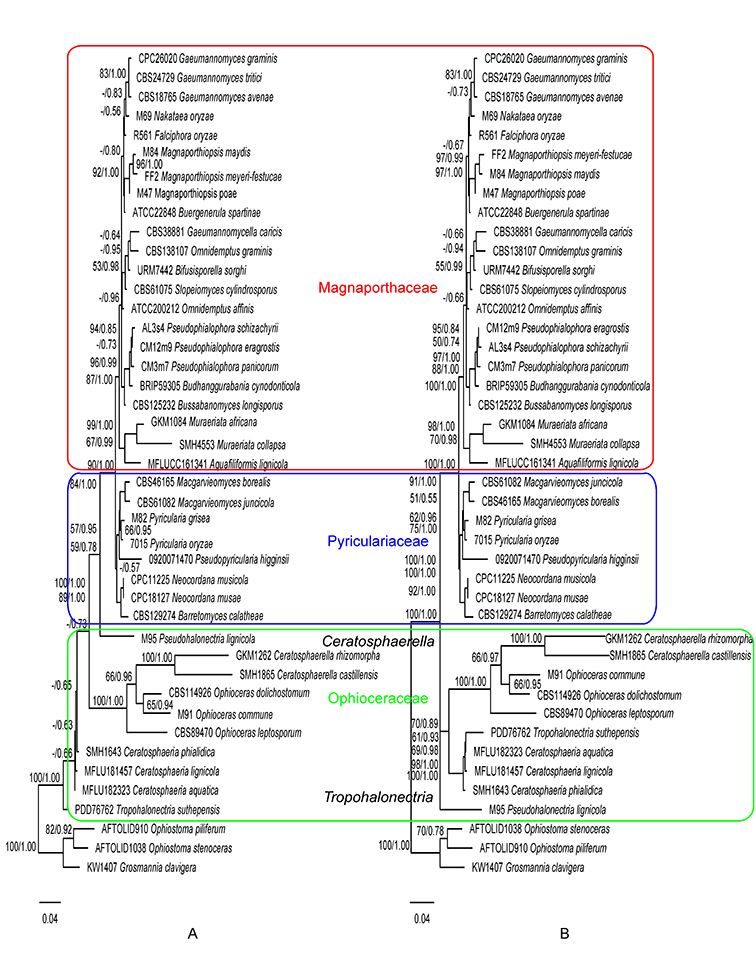 Fig. 1.7. Maximum likelihood trees inferred from the 28S gene sequences without constraint (A), and with Magnaporthaceae, Pyriculariaceae, and Ophioceraceae as constraints (B). ML bootstrap values and BI posterior probabilities are indicated above internodes.
Fig. 1.7. Maximum likelihood trees inferred from the 28S gene sequences without constraint (A), and with Magnaporthaceae, Pyriculariaceae, and Ophioceraceae as constraints (B). ML bootstrap values and BI posterior probabilities are indicated above internodes.
Previously Ceratosphaerella and Muraeriata had been placed in Magnaporthaceae and shared similarities with Ceratosphaeria, Ophioceras, and Pseudohalonectria, including long-necked perithecia, woody habitats and saprobic habit (Huhndorf et al., 2008). However, our results suggested a close relationship between Ophioceras and Ceratosphaerella within the family Ophioceraceae. Muraeriata fell into the family Magnaporthaceae, showing affinity with Aquafiliformis. Unlike other plant pathogenic family members, both genera are saprobic.
Tropohalonectria was proposed for a species that was previously treated as Pseudohalonectria but genetically different from Pseudohalonectria sensu stricto. It was phylogenetically basal in Magnaporthales (Perera et al., 2016). Our study showed similar results and supported its placement in Ophioceraceae.
Phylogenies generated in this study corroborate the three-family system of Magnaporthales according to Klaubauf et al. (2014) and Luo et al. (2015a). Recently the family Ophioceraceae was further split into three families by Hongsanan et al (Hongsanan et al., 2017) and Luo et al. (Luo et al., 2019). However, the two new families Pseudohalonectriaceae and Ceratosphaeriaceae were both based on a single genus and lacked adequate morphological or biological differences to differentiate them at the family level. Therefore, we follow the family circumscription of Ophioceraceae sensu Klaubauf et al. (Klaubauf et al., 2014). In this monograph, the three-family system is adopted, and the placement of genera at the family level is inferred from the ITS, 28S, and TEF1 gene (Fig. 1.6) and the 28S gene only phylogenies (Fig. 1.7).
Copyright 2022 by The American Phytopathological Society. Reproduced, by permission, from Luo, J., and Zhang, N. 2022. The Rice Blast Fungus and Allied Species: A Monograph of the Fungal Order Magnaporthales (https://my.apsnet.org/APSStore/Product-Detail.aspx?WebsiteKey=2661527A-8D44-496C-A730-8CFEB6239BE7&iProductCode=46826). American Phytopathological Society, St. Paul, MN.