Post Views:
1,703
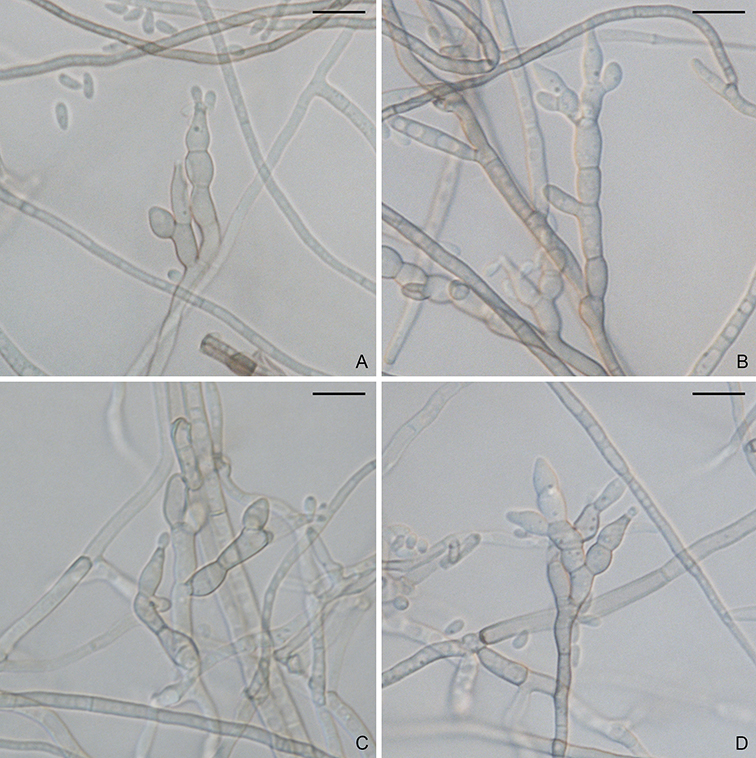
Figure. Magnaporthiopsis meyeri-festucae (FF2). A–D. Conidiophores and conidia. Scale bars: A–D = 10 µm.
Magnaporthiopsis meyeri-festucae J. Luo & N. Zhang.
MycoBank: MB821495.
Asexual state phialophora-like. On CMA, hyphae branched, septate, hyaline to light brown, smooth, 2–4.5 µm diam. Conidiophores macronematous, solitary, erect, straight or curved, branched, brown, smooth. Conidiogenous cells phialidic, light brown, straight or slightly curved, 7.5–14.5 × 2.5–5 µm. Conidia aggregated in slimy heads, ellipsoidal, straight or slightly curved, aseptate, hyaline, smooth, 3–5 × 1–2.5 µm. Sexual state unknown.
Colonies on PDA 4.5 cm diam after 7 days at 25 °C in dark; surface floccose, warbler green; aerial mycelium yellowish to brownish; reverse olive-green. Colonies on CMA reaching 5.3 cm after 7 days in dark at 25 °C; surface sulphur yellow; aerial mycelium sparse; reverse light greenish-yellow (Description from Luo et al., 2017).
Typification: Holotype RUTPPFF2. Ex-holotype culture CBS143324 (FF2).
Gene sequences: MF178140 (18S), MF178146 (ITS), MF178151 (28S), MF178156 (MCM7), MF178162 (RPB1), MF178167 (TEF1).
Materials examined: USA, Maryland, Crofton, from roots of Agrostis (bentgrass)/Poa annua (annual bluegrass) mix, 30 Jul. 2019, S. Tirpak and R. Buckley, RUTPPD30900-1, RUTPPD30901-1; Massachusetts, Amherst, from roots of Festuca rubra var. rubra (strong creeping red fescue), 1 Aug. 2018, Michelle DaCosta, RUTPPscr7-2; ibid., Festuca rubra var. falax (Chewing’s fescue), 1 Aug. 2018, M. DaCosta, RUTPPC39-3, RUTPPC39-6, RUTPPC39-8; New Jersey, Bernardsville, from roots of Poa annua (annual bluegrass) mix, 15 Aug. 2019, S. Tirpak and R. Buckley, D31094-1, Hamburg, from roots of Agrostis (bentgrass)/Poa annua (annual bluegrass) mix, 30 Jul. 2019, S. Tirpak and R. Buckley, D30885-3;ibid., New Brunswick, Hort Farm 2, from roots of Festuca brevipila (‘Beacon’ hard fescue), 26 Jun. 2013, A. Grimshaw, RUTPPFF2, RUTPPAG2); ibid., from roots of Festuca rubra var. rubra (strong creeping red fescue), 17 Jun. 2016, P.L. Vines and L. Hoffman, RUTPPSCR9, RUTPPSCR11, RUTPPSCR12; ibid., from roots of Festuca brevipila (‘Beacon’ hard fescue), 17 Jun. 2016, P.L. Vines and L. Hoffman, RUTPPH11, RUTPPH18, RUTPPH21, RUTPPH31, RUTPPH35, RUTPPH43.
Hosts/substrates: From roots of Agrostis, Festuca, and Poa annua (Poaceae).
Distribution: USA (Maryland, Massachusetts, New Jersey).
Notes: This species causes summer patch disease on fine fescue turfgrasses (Luo et al. 2017).
Copyright 2022 by The American Phytopathological Society. Reproduced, by permission, from Luo, J., and Zhang, N. 2022. The Rice Blast Fungus and Allied Species: A Monograph of the Fungal Order Magnaporthales (https://my.apsnet.org/APSStore/Product-Detail.aspx?WebsiteKey=2661527A-8D44-496C-A730-8CFEB6239BE7&iProductCode=46826). American Phytopathological Society, St. Paul, MN.
